Figure 1 http://blog.neuinfo.org/index.php/news-events/celegansdeepspace
Genetic regulation of organ development and programmed cell death - apoptosis, was discovered in a model organism called C. elegans thanks to the work of John Sulston (UK) and Robert Horvitz (USA). They were both awarded a Nobel Prize in 2002 for physiology and medicine.
Apoptosis is a, evolutionaly conserved programme of cellular self-destruction, which is absolutely essential for development and survival of most multi-cellular organisms.
Figure 1: C.elegans microscope image
C. elegans:[]
C. elegans is a transparent nematode (Figure 1) , which is approximately 1mm long. The first experiments on C.elegans were performed in 1974 by Sydney Brenner and since that time it is a very popular and widely used model organism of molecular and developmental biology.
C.elegans as a model organism :[]
C.elegans is a very advantageous model organism, since it is easily cultivated and sustained (feeds on E. Coli) and its generation period takes only 3,5 days. It has two natural sexes: hermaphrodites (XX) and males (XO), but the absolute majority of the population are hermaphrodites (Figure 2). Hermaphrodites are somatically female, but they can reproduce either by self-fertilization or by mating with males. C. elegans has a simple body consisting of an exact amount of cells. Hermaphrodites (XX) have 959 somatic cells, whereas males (XO) have 1090 cells. C.elegans is also characteristic for having very well differentiated tissues, including the nerveous system. It's body is transparent throughout the whole cycle, which makes it very easy to study under a microscope.
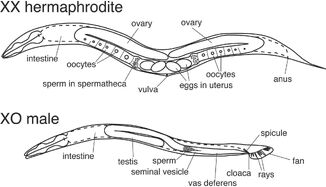
Figure 2: http://www.wormbook.org/chapters/www_somaticsexdeterm/somaticsexdeterm.html
Figure 2: Anatomy and sex determination of C.elegans
C.elegans cell lineage:[]
The C.elegans zygote undergoes a series of asymetric cell divisions which generate six types of blastomeres: AB, MS, E, C, D and P4 (Figure 3). These blastomeres undergo another series of asymmetrical cell divisions - i.e. the blastomere AB is a founder cell for neurons, muscle cells and some epidermal cells.
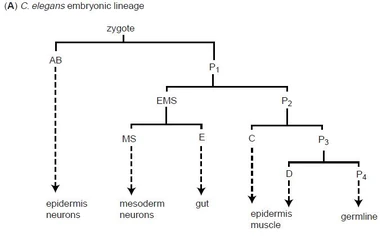
Chisholm A.D., 2001
Figure 3: C.elegans cell lineage
Apoptosis during development:[]
C. Elegans has a invariant and therefore reproducible pattern in every experiment: it is known which cell divides, which undergoes apoptosis and which migrates in that exact time. Out of 1090 cells that are generated to form a hermafrodite, exactly 131 cells always go into apoptosis. If apoptosis wouldn't occur, most of these cells would develop into neurons.
The apoptotic pathway in C. elegans:[]
There are two stages of programmed cell death in the life of C.elegans, which occur in two different types of tissues. The first one occurs during the embryonic and post-embryonic development of the soma (= developmental cell death) and in the gonad of adult hermaphrodites (= germ cell death"). From the 131 cell which die during the development of C.elegans, 113 cells die during during embryonic development and only 18 cells die during post-embryonic development. There were identified over 20 genes involved in the apoptotic pathway of C. elegans. The cell death includes three steps (Figure 4):
- cell decision to die = specification phase
- cell death - activation of the apoptotic program and cell is instructed to die = killing phase
- dead cell recognition by neighbouring cells and phagocytosis = execution phase
- degradation
Figure 4: a microscope image of a cell undergoing apoptosis
The specification phase:[]
Cells in C.elegans are not instructed to die by neighbouring cells, as is normal in higher organisms, but the apoptotic pathway is regulated in a self-dependent manner. The specification whether a cell should live or undergo apoptosis depends on the level of EGL-1 activity. EGL-1 levels are high in cells which are destined to die. The EGL-1 activity appearst to be regulated on the level of transcription.
The killing phase:[]
The essential four genes involved in apoptosis are: ced-3, ced-4, ced-9 and egl-1. Loss of function experiments revealed that ced-3, ced-4 and egl-1 genes are involved in the pro-apoptotic pathway, since all cells survived during their knock-out. Ced-9 on the other hand has an anti-apoptotic effect, since ced-9 gain of function mutants block apoptosis and loss of function mutants are exhibiting excessive cell death. It can be therefore concluded that Ced-9 functions as a regulator, Ced-4 is an adaptor protein of the apoptotic signalling pathway and ced-3 is an effector executing cell death, since it encodes a protease of the caspase family (Figure 5).The egl-1 protein functions as an inhibitor of the ced-9 regulator. Ced-3 and Ced-4 oligomerise together and form an apoptosome-like complex. The process of cell death starts by cleavage of the precursor form of Ced-3: pro-Ced-3, which leads to creation of an active enzyme. Ced-4 is prevented to interact with Ced-3 by direct interaction with Ced-9. This interaction is disrupted by binding of egl-1 which triggers a conformational change of ced-9 that leads to releasing of the ced-9/ced-4 interaction.
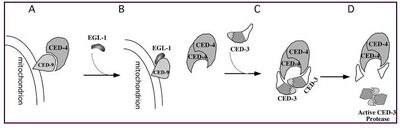
Figure 5: Conradt B., 2005
Figure 5: Biochemical model for the activation of programmed cell death. (A) In normal cell, not undergoing apoptosis, CED-4 is bound to the sufrace of mitochondria by its interaction with CED-9. (B) THe apoptosis activator EGL-1 binds to CED-9, which leads to the release of CED-4 and its successive binding to the pro-caspase CED-3 (C). The procaspase pro CED-3 undergoes an autoproteolytic activation and forms an active executioner caspase CED-3.
Execution phase:[]
This phase involves expression of genes involved in nucleus fragmentation, termination of DNA replication and gene transcription, such as: nuc-1, cps-6, wah-1, crn-1 to 6 and cyp-16. In this phase, apoptotic cells also have to present "eat me" signals on their surface, so that they would be recognized and engulfed by neighbouring cells.
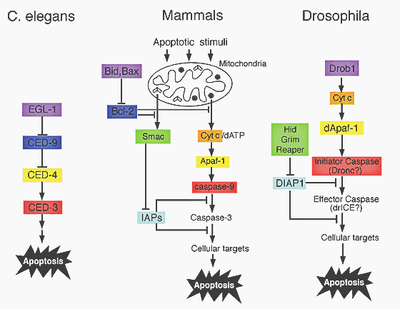
Figure 6 A structural view of mitochondria-mediated apoptosis Yigong Shi
Figure 6: apoptotic pathways are genetically conserved. Proteins that share a homology are displayed in same colors.
Apoptotic pathways are genetically conserved:[]
Apoptotic pathways are genetically very conserved (Figure 6). It has been shown, that Bid and Bcl-2 are highly homologous in sequence and in function to Egl-1 and Ced-9 (Figure 7). Bcl-2 therefore functions as a regulator and Bid/Bax proteins function as regulators of Bcl-2. The mammalian caspase 9 and Apaf-1 also show homology to Ced-4 and Ced-3 in C. elegans. The activation of procaspase-9 is similar to the activation of Ced-3, but there is also a need of Cytochrome C involvement.

Figure 7: Hengartner and Horvitz, 1994
Figure 7: sequence homology between Bcl-2 and CED-9
Experiments used to determine genes of the apoptotic pathway:[]
The first cell death gene that was identified in C.elegans by John Sulston was nuc-1 (nuclease abnormal), which controls the activity of a DNA endonuclease. This gene was discovered by a set of screening experiments. C.elegans was screened for individuals with an abnormal amount of cells in their nerveous system. To visualize these cells, Fleulgen DNA staining was used. It was discovered, that one mutant formed individual DNA positive pycnotic bodies located precisely within positions, where programmed cell death should have occured. This mutant was defective in DNA degradation.
To identify genes, which are responsible for causing cell death, Mr. Sulston and Mr. Brennen sought mutants, which exhibited abnormal cell death patterns. They used nul-1 mutants to visualize cell death and obtained a variety of mutants with abnormalities in patterns of apoptosis. To identify the executive caspase CED-3 they have conducted a mutagenesis of CED-1 defective animals and they found a mutant, in which no dead cell could be found. In a series of experiments it was then proven, that if the CED-3 gene is mutagenized, no cell death occurs in C.elegans.
References:[]
Chicholm A.D., Cell lineage, Academic Press, 2001 Hentgartner M.O and Horvitz H.R. C.elegans Cell Survival Gene CED-9 Encodes a Functional Homolog of the Mammalian Proto-oncogene Bcl-2 , Cell, 76:665-676 (1994)
Horvitz H. R., Worms, life and death, Nobel Lecture, ChemBioChem, 4: 697-711 (2003)
Lettre G. and Hengartner M.O., Developmental apoptosis in C. Elegans: A complex CEDnario , Molecular Cell Biology, 7: 97-108 (2006)
Shi Y., A structural view of mitochondria - mediated apoptosis , Nature Structural Biology, 8: 394-401 (2001)
Zarkower D., Somatic sex determination , WormBook