Theories of Inheritance[]
Prior to the work of Gregor Mendel prominent theories surrounding inheritance, or the passing on of physical traits to one's offspring, involved the idea of "blending." In this type of system, an individuals traits are determined by mixing of the the parental traits into some kind of average value. Strictly speaking, the idea of blending inheritance is not formally a scientific theory as it was never proposed to the scientific community, nor can it be attributed to any single person or group.
Mendel's experiments with peas gave the first concrete foundation for establishing inheritance as particulate rather than acquired, and offered simple mathematical rules for modeling how some traits can be passed from generation to generation. Mendel's work did not gain much attention however until the mid to late 19th century, after his death. With wider acceptance of his work, scientists began to search for the molecules responsible for the heritability of physical traits.
In particular, the chromosome theory of inheritance became a major focal point during the late 19th century. Genetic test-cross experiments with fruit flies by Thomas Hunt Morgan in the early 20th centruy provided solid evidence that chromosmes were indeed the fundamental unit of heritabiltiy. Ironically, Morgan himself had strongly resisted the idea (he was trapped in the pre-Mendelian paradigm of blending) until his experiments with fruit flies showed that the mutation for white eyes followed a sex-linked chromosomal pattern of inheritance.
This was not a complete picture though; chromosmes contain both both protein and DNA components. Many scientists debated the pros and cons of either protein-based or DNA-based theories of inheritance, but it was not until several decades after Morgan's test-cross experiments that more evidence began to accumulate to support a DNA-based theory.
Sources:
http://en.wikipedia.org/wiki/Genetics
http://www.nature.com/scitable/topicpage/thomas-hunt-morgan-and-sex-linkage-452
Proteins vs. DNA[]
The chromosme theory of inheritance marked an important step in the field, but it was still not known whether proteins or DNA are responsible providing the blueprint of an organism. Many prominent scientists argued for proteins, because after all, there were 20 amino acids compared to the meager 4 nitrogenous bases of DNA. A crucial study on the topic was performed by A.D. Hershey and Martha Chase during the early 1950s. They showed that the T2 bacteriophage released it's DNA into a bacterial host and not it's protein coat in an elegantly simple series of experiments, described below.
Hershey and Chase: Experimental Setup and Technique[]
Hershey and Chase based their experiments on two basic yet very important attributes of proteins vs. DNA: that DNA contains phosphorus while proteins contain sulfur. Knowing this, they were able to differentially radiolabel each macromolecular component: P-32 was incorporated into DNA while S-35 was incorporated into the virus's protein. These radioactive isotopes were added to the bacterial growth media, which was then infected with the T2 phage. After several hours the phages had assimilated the different isotopes, and they were seperated

General outline of the Hershey & Chase experiments. Source: http://en.wikipedia.org/wiki/File:Hershey_Chase_experiment.png
from the bacteria by several cycles of slow (<2000 x g) to fast (12,000 x g) (centrifugation.
Once they had a supply of radiolabeled T2(note that each phage had either P-32 or S-35, not both), they performed numerous assays aimed at detecing the radioactive isotopes following adsorption of the phages to bacteria grown in non-radioactive media. After allowing time for the phages to adsorb to the bacteria, the researchers would put the solution in a blender which provided enough force to dislodge any phages attached to the surface. Following this, centrifugation at high speeds pelleted the bateria while the phage capsules remained in solution.
Results[]
Hershey and Chase noted many important findings in their paper following these experiments. Notably, after adsorption of the phage to bacteria and subsequent seperation by centrifugation, the bacterial cells were lysed and P-32 DNA was released into solution. Furthermore, approximately 80% of the phage sulfur could be easily stripped from the bacteria by agitation, and it remained in the solution phase following centrifugation. Conversely, only 21 to 35% of the radioactive phage phosphorus could be found in the solution and not the pellet.
Hershey and Chase showed that when a T2 phage infects bacteria, the majority of it's DNA enters the bacterial cell. The sulfurous protein on the other hand remains on the cell surface, signifying that it does not play a role in infecting the bacteria outside of inital attachment of the phage. Altogehter, this landmark paper provided the strongest argument for a DNA-based theory of inheritance.
For an animation outlining the Hershey-Chase experiments, try this.
Source:
Hershey A, Chase M (1952). "Independent functions of viral protein and nucleic acid in growth of bacteriophage" (PDF). J Gen Physiol 36 (1): 39–56. doi:10.1085/jgp.36.1.39. PMC 2147348. PMID 12981234. http://www.jgp.org/cgi/reprint/36/1/39.pdf.
New Techniques, New Findings[]
Fast forward only a few years from the Hershey-Chase experiments and it is already unequivocaly accepted that DNA is the molecular component responsible for heritability. Technological advances during this time opened up a whole vista of possibiltiies. In a paper by Tsugita et al., (his lab will re-appear later in this discussion) his group solved the complete amino acid sequence of the subunit of the Tobacco Mosaic Virus (TMV). The techniques they used provide a great overview of the technological system utilized during this time period for protein-sequencing endeavors.
Many of the earliest sequencing projects focused on the TMV, primarily due to the ease at which it can be obtained and purified. Furthermore the protein coat of the virus is a repeating peptide, which at the time was relatively easy to work with because of it's small size and large quantities (the capsid is now known to be made up of 2,130 of these peptides).
To sequence small peptides, early researchers used a series of sequential digests that would chop up the protein into smaller pieces that could be carefully analyzed to improve accuracy of the read. This was usually initiated by either a trypsin or chymotrypsin digest. Trypsin and chymotrypsin are proteases that cut at specific locations within a polypeptide chain. Trypsin cleaves peptides on the carboxyl-end of either lysine or arginine, except when the next C-terminal amino acid is proline. Chymotrypsin cleaves peptides on the carboxyl-end of either tyrosin, phenylalanine, or tryptophan. By digesting proteins with these enzymes, more manageable pieces can be acquired. Techniques such as these remain widely used today.
Following these initial digests, the peptide fragments can then be subjected to seperation techniques such as paper chromatography and electrophoresis. In the Tsugita paper, some of his analyses involved the digestion of the peptides with another protease, called nagase (subtilisin). Another technique that was just begin
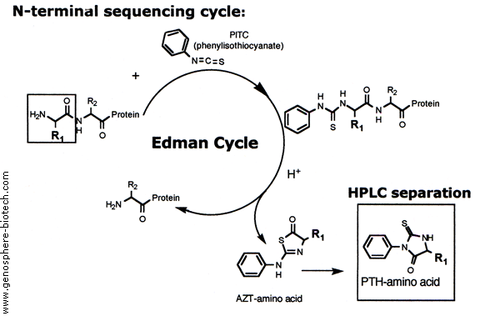
Outline of one Edman cycle in an Edman degradation experiment. Source: http://www.genosphere-biotech.com/Edman-Degradation.html
ning to be used by the scientific community was the Edman degradation (also known as N-Terminal sequencing), pioneered by Pehr Victor Edman in 1950. This method quickly rose to critical importance for protein sequencing: it allowed a N-terminal amino acid (uncharged) to be cleaved off of a peptide without disrupting the other bonds. Subsequent seperations techniques, most often high performance liquid chromatography, can be used to identify the released amino acid after being compared to profiles of known standards. This method has strict limitations however, as the reaction does not always go to completetion and side reactions produce intermediates and by-products that obscure results after multiple cycles. This is the primary reason for the initial protease treatments.
Sources:
- Tsugita et al. (1960) PMID
- 16590772
http://en.wikipedia.org/wiki/Tobacco_mosaic_virus
http://www.genosphere-biotech.com/Edman-Degradation.html
Deciphering the Code[]
While this was a large step in the right direction, many questions still arose regarding the genetic code. It was obvious that nucleotides did not code for amino acids in a 1:1 ratio; if that were the case there would only be four possible amino acids. The code could not be 2:1 either, as this only gave rise to 16 possibilities. The smallest possible number would have to be three, a triplicate code. What about spacing though? Did every nucleotide participate in coding, or do some act as "spacers" or commas? Was each sequence of three (or more) nucleotides itself a discreet element , or did the code overlap?
Some of the first initial evidence against an overlapping code came out of Tsugita's lab in Berkeley, California. After solving the sequence of the TMV coat protein (mentioned earlier), his group began work and eventually published a paper in 1960 surrounding the effects mutations in DNA had on the resulting protein.
They began by performing phenol extractions on the nuclear material (in this case it was actually RNA, not DNA). This is a commonly used technique that seperates DNA and RNA from proteins. Phenol is less polar than water, and DNA is not soluble in this type of solvent. Proteins on the other hand are still soluble in an organic solvent such as phenol (note that they are usually irreversibly denatured however). Thus this mehtod purifies nucleic acids from proteins while also protecting them by denaturing any DN or RNAses.
Following this, the RNA was subjected to various mutagens including nitrous acid, dimethyl sulfate and N-bromosuccinimide. In particular, nitrous acid performs a nucleophillic attack on certain bases and causes de-amination. When this was complete the RNA was re-precipitated with another phenol extraction and reconstituted with viral protein.
To test for mutations, the reconstituted virus/RNA was applied to plants that the TMV is known to infect (Java Tobacco and N. sylvestris) and then the plants were monitored for abnormal lesions and growths. The "wild-type" virus (that which had not been exposed to mutagens) rarely caused lesions, while mutated ones did at a 100-fold increase in occurence. (Unfortunately, none of this data was shown in the paper)
Overall, hundreds of mutant TMV strains were then isolated from these lesions and their protein content was analyzed using similar procedures discussed earlier (protease digests, etc.) with the addition of one of the first automated amino acid anlyzers, which became commercially available 2 years before this paper was

Overlapping code compared to a non-overlapping code. Source: http://www.nature.com/scitable/topicpage/reading-the-genetic-code-1042
published.
Interestingly, it was determined that a single point mutation had occured in several of their TMV mutants. After sequencing the resulting protein from these mutants, they observed that only a single amino acid had changed. This was a striking discovery, as it strongly implied that the DNA code could not be overlapping. If the code overlapped, a single mutation would cause a change in more than just one amino acid; as multiple codons would be affected.
Sources:
- Tsugita et al., 1960. PMID: 16590652
The Possibility of a Triplicate Code[]
Initial Work[]
The genetic code was brought into even more light by the Dr. Francis H. Crick less than a year after Tsugita's second publication. Crick and coworkers devised a clever system using frameshift mutations in the T4 bacteriophage and analyzing the virus's ability to grown on types of bacteria.
To induce frameshift mutations, the viral DNA was exposed to acridines such as proflavin. Proflavin is known to produce indels within a DNA sequence. Dozens upon dozens of mutants produced this way were then screened to find those that had either a single basepair insertion or deletion. The mutated virus's ability to infect certain species of E.coli was then assesed. Before explaining the results, first consider the experimental setup:
1) Wild type T4 can infect both K12 and B strains of E.coli.
2)T4 has two cistrons, both of which must be intact and functional for it to infect K12. Mutant T4s that have lost funcitonality in either or both genes will not be able to infect K12, and will grow a characteristic plaque on the B strain.
3)The goal is to mutate the DNA in these cistrons, as the integrity of these regions does not appear to be essential for phage survivability.
4)It is not known (at the time...) whether these cistrons encode for proteins. Instead of looking at amino acid composition, they simply assesed the phage's ability to infect the different strains of E.coli.
Crick and colleagues hypothesized that if their acridine treatment deleted or added a base at one location, a second treatment that added or deleted, respectively, could restore the reading frame and result in a reversion back to a funcitoning protein. They tested their hypothesis by screening for these secondary "rescue" mutations and culturing the virus on both strands of bateria.
Coincidently, secondary mutations that restored the correct number of nucletotides (i.e., an addition followed by a deletion, or vice versa) resulted in phages that were indeed able to infect the bacteria. That is not to say that the secondary mutation returned the phage to a wild-type genotype status, but at the very least restored a wild-type phenotype by correcting the reading frame.
Triple Mutants[]
Now this alone was not enough to claim that DNA is a triplicate code. To do this the researchers performed further experimentation on acridine-induced T4 mutants that had either lost three bases or gained three; triple mutants (in practice only triple addition mutants were produced). Once these mutants were identified, it was shown that they could, in some cases, infect the K12 strain.
Upon further analysis, they realized that if the three additions were close together within the same cistron, the phage was able to infect K12. However, if the additions were seperate and located within a different cistron then the phage lost its ability to infect K12. It thus appeared that only mutations that resulted in either 1 or 2 additions/deletions caused a loss of virulence for T4. When three mutations occured all together, the virus retained it's infectability.
This presented an extremely strong case for a triplicate code that did not overlap and was not punctuated. Strictly speaking however, it did not necessarily rule out the possibilty that the genetic code is read in ratios of 6, or another multiple of 3. The nature of their screening for single addition/deletion mutatnts is not perfect; there is always the possibility of error. However their paper was the strongest case for a triplicate code at the time and represented an incredible leap forward for the field of genetics.
Sources:
Crick et al., 1961. PMID: 13882203