Challenges to the Central Dogma[]
The central dogma of molecular biology is that DNA is transcribed into RNA, which is then translated into proteins.

Illustration of the central dogma of molecular biology. Source: http://en.wikipedia.org/wiki/Central_dogma_of_molecular_biology
However, recent discoveries of alternative roles played by various RNAs have begun to challenge the universal nature of this paradigm. Thousands of large RNA molecules have been identified that do not serve as templates for protein synthesis; rather, they carry out functions relating to epigenetic regulation at the transcriptional, post-transcriptional, and even translational levels. While much of the world of these long, non-coding RNAs (lncRNAs) remains a scientific enigma, recent publications have begun to enhance our understanding of the various roles they play.
Characteristics of lncRNAs[]
As an operational definition, lncRNAs are RNA genes that are at least 200 bps long and have no protein-coding potential. While the restriction on size is somewhat arbitrary, it distinguishes them from the numerous small, regulatory RNA sequences such as microRNAs, snoRNAs, and piRNAs. Various transcriptomic studies have indicated that lncRNAs make up a significant portion of mammalian genomes, providing evidence against the idea that much of an organism's genome (especially humans) contains "junk DNA".
Biochemically speaking, lncRNAs are similar to mRNA transcripts; they are post-transcriptionally processed by addition of a 5' mehtyl-guanine cap and undergo polyadenylation on their 3' ends. At the gene level the vast majority are spliced; containing flanking splice sites at their 3' and 5' ends, and about 25% have alternatively spliced variants. In contrast to most protein encoding genes, lncRNAs are (for the most part) poorly conserved and have a high degree of variability between species as well as individuals. This lessened evolutionary pressure has led to the hypothesis that only smaller sequences within the lncRNA transcript itself are required for structure or sequence-specific interactions relating to their function.
Similar trends have been observed across the kingdoms of life, although much research has gone into murine lncRNAs. It has been shown in mice that lncRNAs are comparable in overall stability and half-life duration to mRNA transcripts (likely due to the extent of post-transcriptional modifications), although they are often expressed at much lower levels which makes them difficult to detect and annotate.

Possible mechanisms of lncRNA action by formation of RNPs. Source: Rinn, J.L. & Chang H.Y. Annu. Rev. Biochem. (2012) 81:145-166
Lots of evidence has been found that implicates lncRNA functionality to correspond with the formation of ribonucleoproteins with DNA binding proteins such as transcription factors or chromatin remodeling enzymes. As illustrated by the figure to the right, this could be exhibited by a) the lncRNA acting as a decoy to titrate away DNA-binding proteins, b) acting as a scaffold to bring proteins into closer proximity, c) acting as a guide by either recruiting protein to the DNA or DNA to the protein, or d) acting as a guide by creating a chromosomal loop as seen in enhancers.
Xist: Dosage compensation through X chromosome inactivation[]
X-chromosome inactivation is a mechanism of dosage compensation that has evolved in mammals to ensure equal expression levels of genes located on the X chromosme between males and females. The idea that this process occurs through the inactivation of one copy of the X chromosome in females was first proposed by Mary Lyon in 1961. In mice, this process begin during early embryogenesis when the Xist gene is transcriped to produce a 15 kilobase transcript that is localized to the nucleus. This gene is found on the X chromosome in a region called the "X inactivation center" (XIC), which contains gene clusters and regulatory sequences necessary for proper inactivation. Interestingly, Xist expression only occurs on the chromosome that will become silenced.
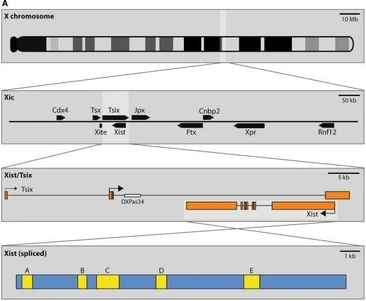
Overview of the XIC, located on the X-chromosome. Source: Pontier, D.B., & Gribnau, J. Hum. Genet. (2011) 130:223-236
As the Xist transcript is produced it begins to coat the X chromosome in cis, forming a nuclear compartment that excludes RNA polymerase III and recruits various chromatin remodeling protein complexes such as PRC2, which modifies the trimethylates histone H3. Methylation of this histone is a characteristic mark of trancriptionally inactive chromatin. X chromosme inactivation occurs in all differentiated somatic cells in mice. Once it has occured it cannot be reversed and is therefore tightly regulated in mice by the antisense Xist lncRNA, Tsix. Numerous other proteins are thought to be involved in the regulation of this process, but mechanisms are still currently uknown.
Several key questions still remain unanswered regarding Xist. One of these questions is how cells are able to tightly localize Xist to only the X chromosome that is going to be inactivated. Various studies have shown that the transcript never leaves the immediate vicinity of the chromosome it originated from. Another is how the Xist transcript recognizes the X chromosome; to date no sequence has been identified that designates the X chromosome for inactivation.
Malat1: Regulation of adjacent genes[]
One of the most highly expressed and well conserved lncRNAs in the mouse genome is Malat1, located on chromosme 19. This molecule is processed after to ~6,700 nucleotides after being transcribed and is strictly localized to the nucleus. It has been shown to have consistently high levels of expression throughout the pre- and post-natal mouse developmental cycles, and it lacks any annotated repeat-derived sequences. Researchers originally saw these characteristics as implying a crucial biological function for Malat1, possibly during early development.
Ironically, experiments with mice having this particular RNA gene knocked out showed that this was not really the case. The loss-of-function mutation was not embryonically lethal; in fact it did not have any detectable effects during both the pre- and post-natal development phases. However, researchers did notice that during the adult mice had statistically significant differences in expression levels for 12 genes either adjacent or in close proximity to Malat1 on chromosome 19.
These changes occured in a tissue-specific manner. For instance, Neat1 (interestingly enough another lncRNA) was found to be upregulated up to 2-fold in the liver and brain cortex. 11 other genes were found to be up-regulated between 1.5 and 2.5-fold. Despite being "dispensable" for development the changes of gene expression observed in adult mice along with the unique characteristics of Malat1 hint that this lncRNA has a broader biological function, possibly being hidden by compensatory or redundant mechanisms.
Sources[]
Clark, MB et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. (2012) 22:885-898
Derrien, T., Johnson, R., and Bussotti, G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution and expression. Genome Res. (2012) 22:1775-1789
Pontier, D. B. and Gribnau, J. Xist regulation and function eXplored. Hum. Genet. (2011) 130:223-236
Rinn, J.L., and Chang H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. (2012) 81:145-166
Zhang, B. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Reports. (2012) 2:111-123